Classifying colorectal cancer with the oral (and gut) microbiome
Flemer et al. 2017. The oral microbiota in colorectal cancer is distinctive and predictive. Gut.
Summarized from DalMUG group discussion and written by: Casey Jones
Summary
The microbiome has been connected to several human cancers, namely those involving the gastrointestinal tract. Specifically in colorectal cancer (CRC), changes in both fecal and mucosal microbiome have been reported and theorized to play a role in CRC pathogenesis (shown recently here).
You may think it’s odd for the oral microbiome to be involved in an intestinal pathology. However, it’s previously been shown that microbes that are normally associated with the mouth find their way into the gut, specifically at cancerous sites in the colon.
In a recent study published in Gut, Paul O’Toole’s group from the University of Cork present a novel microbiome classifier using two previously uncombined data sets - the oral and stool-derived gut microbiota. They studied three groups of patients: controls, CRC patients, and individuals that presented with colonic polyps.
Their major finding is that combining the oral microbiome and stool microbiome datasets distinguished those with cancer and those with colonic polyps compared to controls with a relatively high specificity. They also found that some of these oral microbes are enriched in pediatric Crohn’s disease biopsies, by doing a meta-analysis with a large 2014 study.
This novel classifier has the potential to be used as a non-invasive biomarker for risk of cancer development, say the authors. We found it interesting that the oral microbiome could be used to predict pathologies such as colorectal cancer, and we think that the study’s outcomes are promising for future early-detection methods.
See our main takeaways from the paper below.
Points of Interest
- A novel combination of oral and fecal microbiome datasets improved sensitivity in distinguishing disease. The term “sensitivity” in a random forests model measures the proportion of positives that are correctly identified (i.e. true positive discovery rate). See our summary table here:
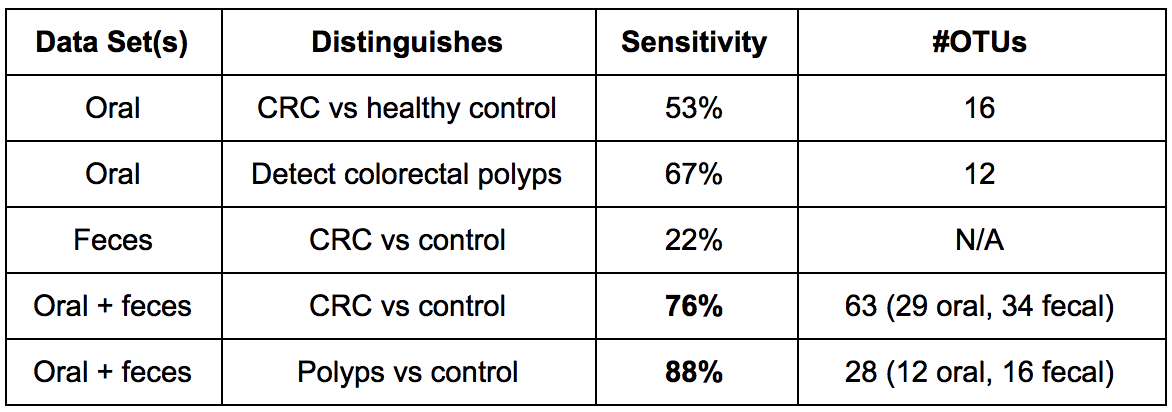
-
Oral pathogens that were previously linked with oral biofilms and human disease (7 OTUs), and dominant bacteria in early dental biofilm formation (10 OTUs) were found to be significantly more abundant in colonic microbiota of CRC patients.
-
They found that colonization of human CRCs with oral bacteria was negatively associated with the presence of Lachnospiraceae in the colonic mucosa, suggesting that this microbe may be protective.
Points of Confusion
- The overlap of the swab, stool and mucosal biopsy samples between patients was unclear (Table 1).
- It was also unclear why these sample groupings weren’t balanced - were the samples grouped retroactively? Furthermore, were the on/off samples from the same patients or not? It doesn’t seem so based off the sample sizes.
- Although controls did not have CRC, IBD, or IBS they still had high percentages of rectal bleeding (up to 58%), suggesting that they are not ideal healthy controls.
- Regarding the random forest methods, could running LASSO for feature selection before running random forests be resulting in overfitting of the model?
- Usage of weighted and unweighted UniFrac distance matrices instead of only unweighted, to compare oral microbiome dissimilarity in each of the three groups studied may have been beneficial. (Figure 1)
- It was unclear as to which oral OTUs were being correlated with the CD samples - was it those from controls, CRC, polyps, or all groups? (Figure 6)
- The association with “Western diet” was very weak and maybe doesn’t merit a mention of such in the Key Messages box.
